Shay “Shai” Maor is a Senior Level Developer with over 12 years of Information Technology experience. He has 10+ years of software development experience including over 10 years Hands on Industry Experience and 6 months Academia Development experience as a Member of a Center for Computational and Integrative Biology Research Group. He has experience in Microsoft C#/.NET (8+ years), Python (2+ years), and Java (1 year). He graduated from Rutgers University in May 2020 with a Bachelor of Science in Computer Science and a Minor in Psychology with a focus on Software Development electives and 2 Computational Biology Independent Studies. He graduated with a Master of Science in Software Development from Boston University (ranked #8 in Online Information Technology Programs by US News) taught by MET Computer Science Faculty with a focus on BU MET Computer Science Electives in 2025.
Content Management Systems
Content Management Systems (CMS): These systems allow content authors to publish content to their website while the developer creates the foundation and features for the content authors. Creating a separation of responsibilities so that authors can focus on creating engaging content and engage with developers if they need custom functionalities added to their websites.
Content Management Systems Experience
Highlighted work includes Search Engines, Commerce Functionality including Wishlist and SEO Metadata, Reporting Automation, Conference Reporting, Responsive Data Visualizations, Custom Content Migrations of over 20,000 articles between different CMS, Custom Webhook APIs, Responsive Upgrades, and much more.
Portfolio
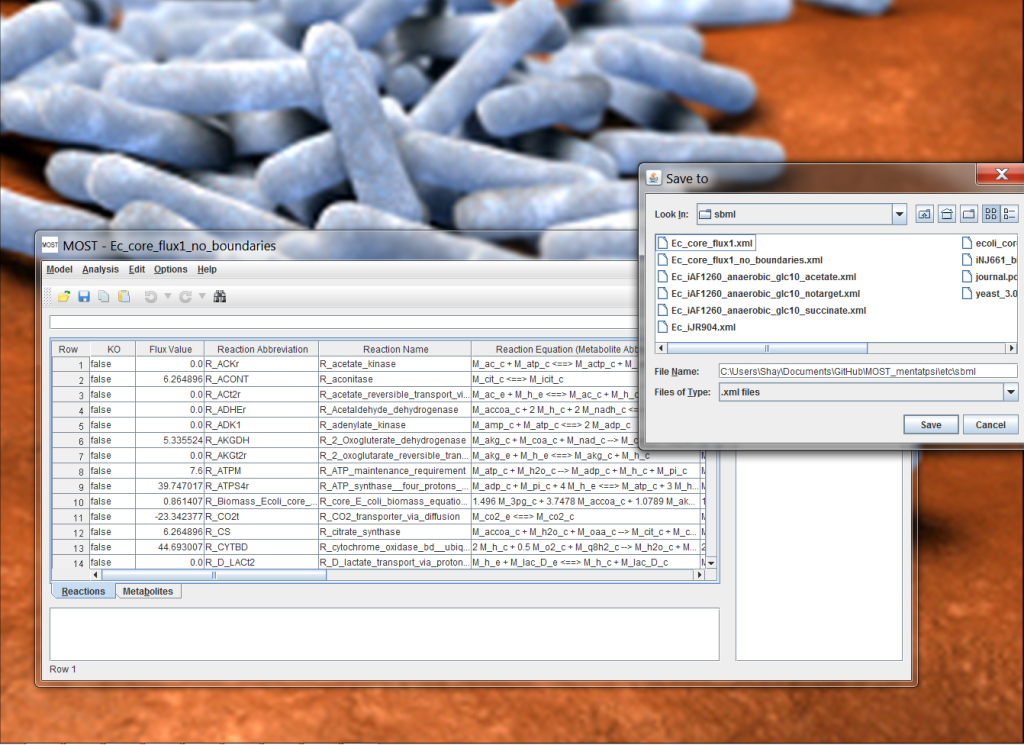
Java based systems biology tool designed to research organisms and their respective metabolic networks, with the intention of both simulation and discovering ideal gene knock-out combinations for increasing production of target metabolites. Originally as part of a Computer Science independent study. Added several components to pre-existing software package.
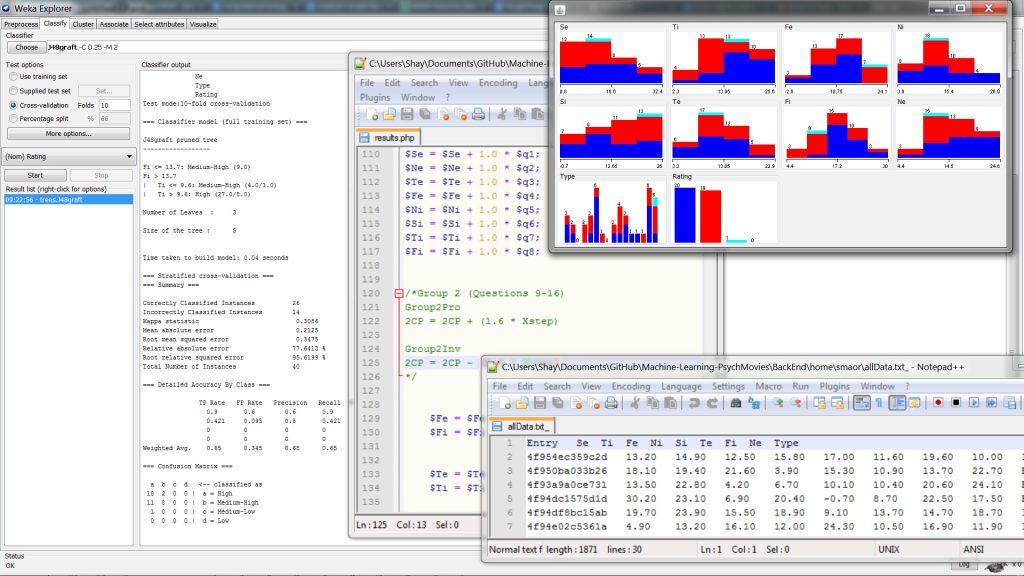
Senior design project that implemented data mining techniques and a psychological assessment test (MBTI) to discover correlations and predictive models on movie preferences. Primarily, using numerical quantification of the 16 cognitive functions underlying the MBTI to find correlation with sub-genres using a J48 decision tree-inducing algorithm under Weka Machine Learning software. The J48 algorithm is Weka’s implementation of the C4.5 algorithm by Ross Quinlan.
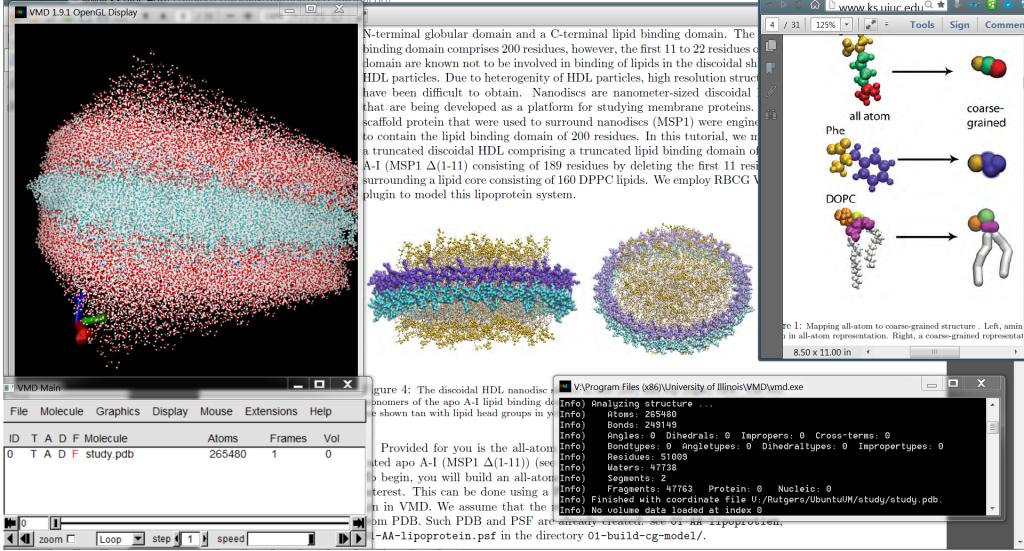
Under a Physics Independent Study, worked on simulating and analyzing coarse grain models of lipid bilayer membranes using Molecular Dynamics software packages (GROMACS, NAMD, VMD) and TCL. The main focus was the recreation of the simulation in the study “Phase Separation in a Lipid/Cholesterol System: Comparison of Coarse-Grained and United-Atom Simulations.”
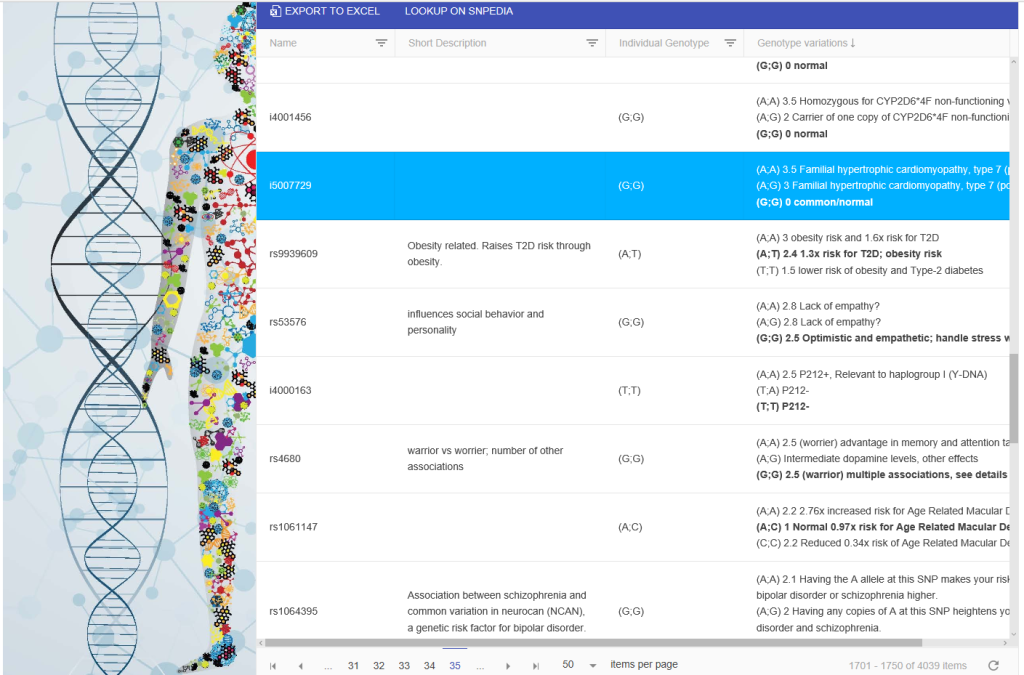
OS Genome is an open source web application that allows users to gather the information they need to make sense of their own genome without needing to rely on outside services with unknown privacy policies. OS Genome’s goal is to crawl various sources and give meaning to an individual’s genome. It creates a Responsive Grid of the user’s specific genome. This allows for everything from filtering to excel exporting.
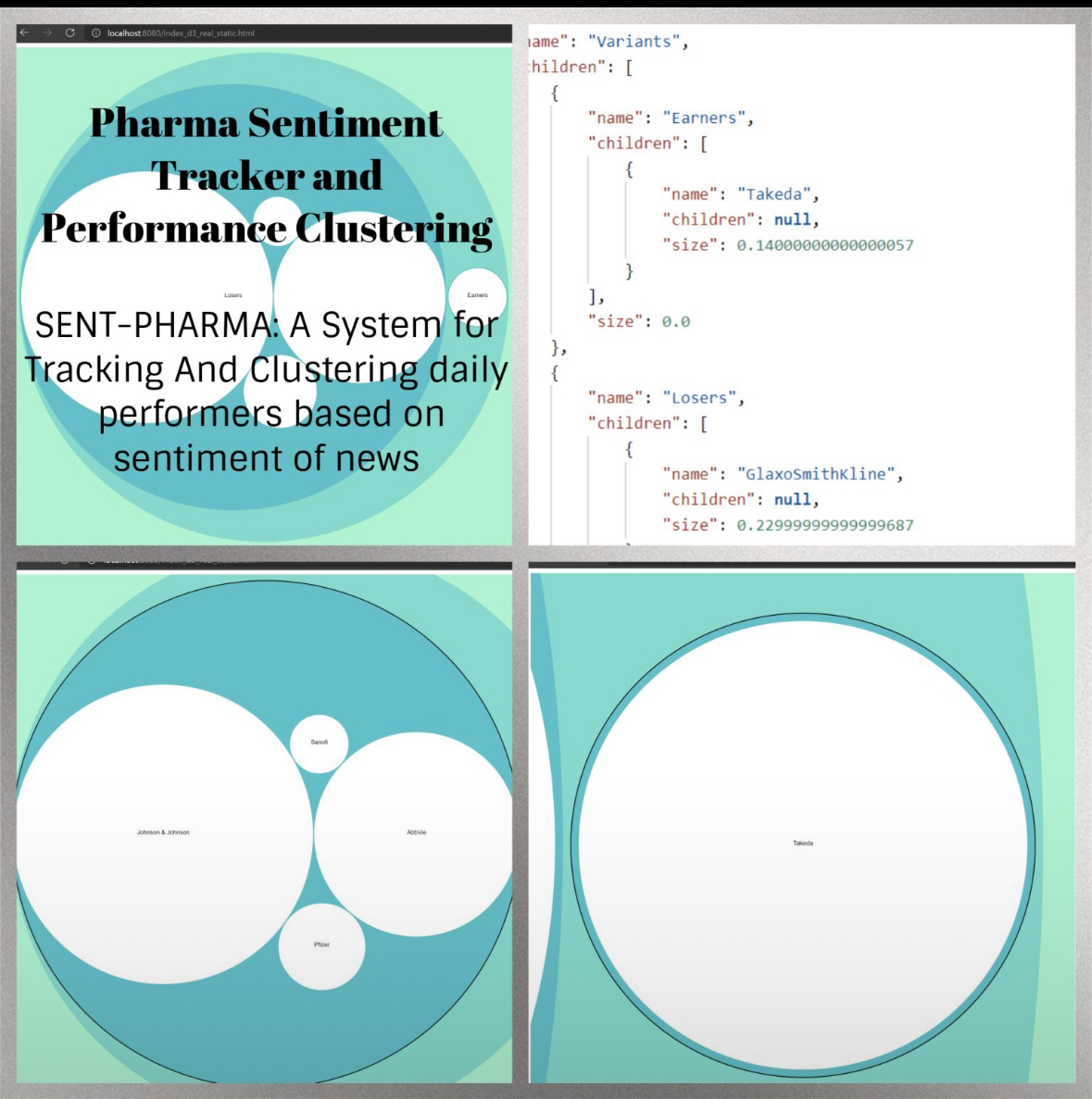
Concurrent Pharmaceutical Sentiment Analysis + Stock Valuation Tracker in Java Spring for my Boston University class, Advanced Programming Techniques. With several key MVC Endpoints for news aggregation, sentiment analysis using Microsoft Azure Cognitive Services, and Stock Valuation time series retrieval. Pictured above is the clustering D3.js data visualization using an MVC Endpoint for data population.
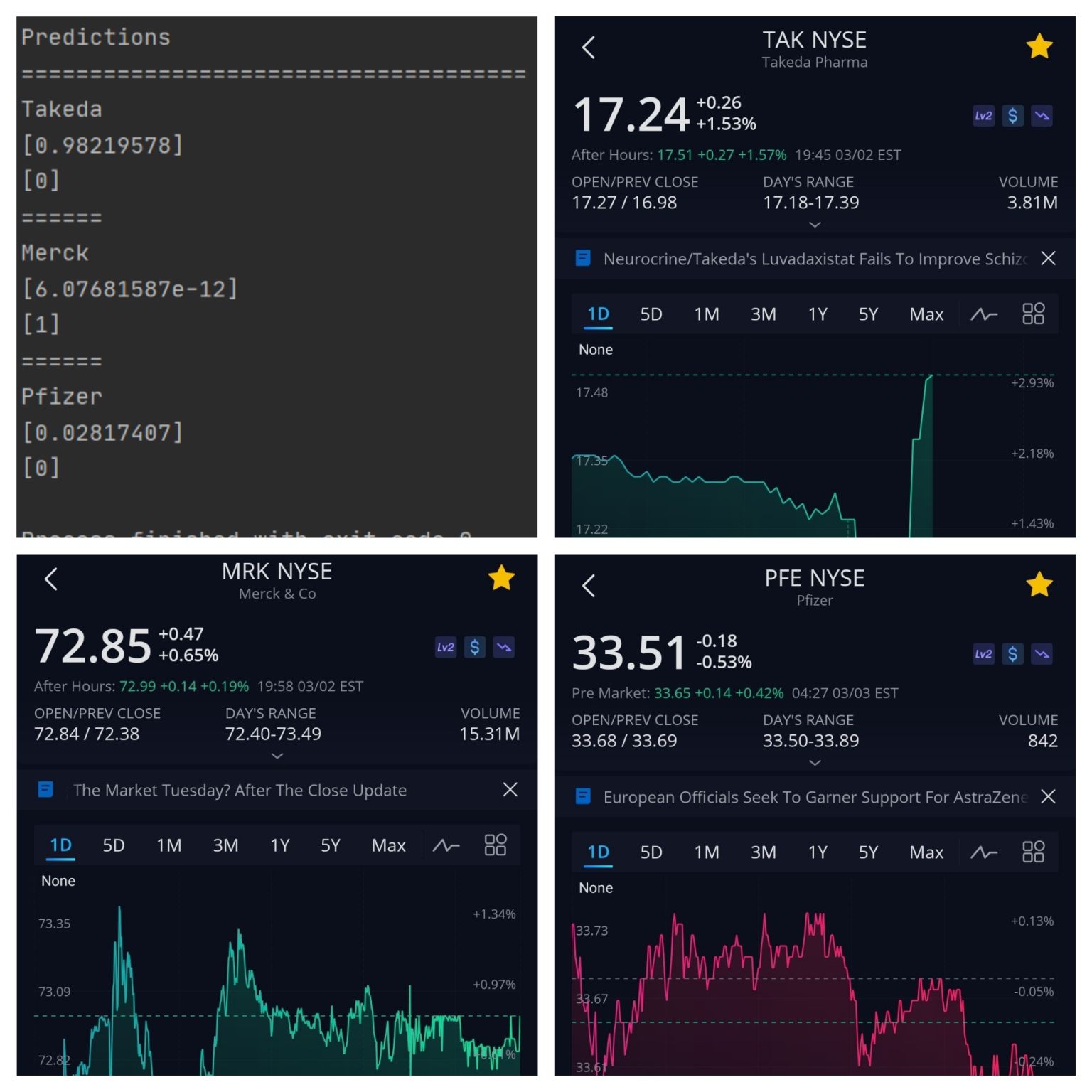
Python script leveraging SQL Alchemy to aggregate and analyze data from the SQL Server I set up on AWS Linux Cloud and a Neural Network using Linear Regression and Sigmoid Function to predict stock market behaviors. Pictured above is the Prediction Algorithm predicting loses for Takeda and Pfizer, and gains for Merck. Takeda had losses after hours, with the prediction accounting for news aggregated after market hours
Highlighted Experience
Recent Experience
Cobblestone Software / Remote / may 2025 – September 2025
Full Stack Generative AI, Natural Language Processing, and other Artificial Intelligence Technique Development for Legal Contracts
Delivered Generative AI Large Language Model Engine with NLP analyzing and processing Legal documents featuring OpenAI and Mistral featuring SignalR, Telerik, and various Full Stack Technologies in .NET
Ad-Hoc On Support Client bug handling
Assisted in Local Environment DevOps with PowerShell script for SQL
Assisted in helping Developers with Environment set up
PJM Interconnection / Remote / Jan 2022 – JAN 2025
Sitecore .NET Full Stack Development under contract for Utility/Energy company
Full Stack Sitecore 9.3 and 10 .NET Content Management System Development.
• Helping migrate Legacy Code to modern MVC architecture pseudo N-Tier Architecture (Sitecore Helix Architecture)
• Mentoring Developers (I and II) in development including code review and pair programming
• Helped migrate from On Premise Hosting to Sitecore Managed Cloud (Azure PaaS)
• Coveo Cloud Search Development
𝐓𝐞𝐜𝐡𝐧𝐨𝐥𝐨𝐠𝐢𝐞𝐬
Sitecore 9.3 CMS, Sitecore Helix Architecture, Microsoft SQL Server, T-SQL, PowerShell, ASP.NET MVC, C#
Caleres, Inc / Remote / May 2021 – December 2021
Parent Company of Famous Footwear, Naturalizer, Sam Edelman, Allen Edmonds, and many more
Sitecore .NET Content Management System, used by Volvo, Merck, PSE&G, Zurich, Cirque du Soleil, and many more
Omni Channel eCommerce Development
– Sitecore Experience Platform (XP)
– Sitecore Experience Accelerator (SXA)
– Sitecore Commerce
- Collaborated with team members to develop a Wishlist Shopping feature, a high value Omni Channel feature across multiple sales channels following Helix Sitecore Design Principles and Architecture
- Implemented modifications to Meta Data for Search Engines by following Helix Model and Leveraging Overrides on Sitecore code
- Participated in quick delivery of feature
- Performing On-Call Rotations examining New Relic Production Alerts
Information Today, Inc / Medford, NJ/ Feb 2019-May 2021
.NET Web Development of Proprietary Content Management Systems (CMS) and Email Blast System.
- Upgraded web magazines (Knowledge Management World, Speech Technology Magazine) to new Responsive Design as .NET Framework CMS Applications utilizing ORM, C#, T-SQL, JavaScript, jQuery, and more
- Data Visualization Feature for Newsletter Advertisement Performance
- Converted Java Tomcat Reporting App to .NET DLL providing access to around 20 On Demand Reports for Conference Management System
- Designed ASP.NET REST API Endpoints and a Web Form for Facial Detection for media creation through Azure Cognitive Services. Attaching Amazon S3 and our proprietary CMS’ binary image data for image stream
Tim Corp of NJ/ Cherry Hill, NJ/ Aug 2017 – Jan 2019
Automation of Reporting, Operations, E-commerce, and Data Engineering using Python, C#, PHP, and PowerShell and Magento CMS
- Created Python Web Scraper to gather Lowest eBay prices based on UPC or Model with fuzzy matching reaching 93% Accuracy
- Lead external Omnichannel e-Commerce Magento CMS Development
Fuel 180/ Remote/ Sept 2015 – Aug 2017
Sitecore 7 development, C#, .NET, jQuery, JavaScript, UI Design.
- Developed Search Website for user inquiry allowing for filtering and searching of products hosted on Sitecore CMS through a data set of product descriptors and metadata. Utilized JavaScript, jQuery, Sitecore 7, C#, .NET, and HTML.
- Ad-Hoc Diagnostic and Troubleshooting of Sitecore websites.
Cubesmart / Malvern, PA / Dec 2016 – Mar 2017
- Built out parallel C# Analysis Application analyzing over a year of financial SQL data for research into justifying alternate vendor payment sytstem
- Built C# Application utilizing Azure Jobs for Data Migration of Azure Blob Storage to MongoDB
Rutgers University/ Camden, NJ / Sept 2012 – JunE 2016
- Worked with team members, and under the guidance of two professors, on a Java based Metabolic Optimization and Simulation Tool designed to research organisms and their respective metabolic networks, with the intention of both simulation and discovering ideal gene knock-out combinations for increasing production of target metabolites through genetic engineering.
- Worked on adding save functionality (JSBMLWriter) to SBML file format (XML) as well as several enhancements discovered during production.
- Added a Settings Factory to allow for session memory using XML.
- Created KEGG-Crawler, an open source Parallel/Multithreaded Python Data Crawler of the Kyoto Encyclopedia of Genes and Genomes API; generating CSVs of Genomic and Metabolic Data. KEGG-Crawler was used to populate the database of MOST-Visualization, a feature providing visualization of metabolites, reactions and pathways in organisms’ metabolic networks.
More information: most.ccib.rutgers.edu
Exposure to Eclipse, GitHub, Java, JSBML, SBML, SQLite, XML, among other languages and software.
Publications
MOST – Visualization: Software for producing automated textbook-style maps of genome-scale metabolic networks
April 2017 Bioinformatics, Oxford Journals
Visualization of metabolites, reactions and pathways in genome-scale metabolic networks (GEMs) can assist in understanding cellular metabolism. Three attributes are desirable in software used for visualizing GEMs: 1. automation, since GEMs can be quite large; 2. production of understandable maps that provide ease in identification of pathways, reactions, and metabolites; and 3. visualization of the entire network to show how pathways are interconnected. No software currently exists for visualizing GEMs that satisfies all three characteristics, but MOST-Visualization, an extension of the software package MOST (Metabolic Optimization and Simulation Tool), satisfies (1), and by using a pre-drawn overview map of metabolism based on the Roche map satisfies (2) and comes close to satisfying (3).
MOST: A software environment for constraint-based metabolic modeling and strain design
October 14, 2014 Bioinformatics, Oxford Journals
MOST (Metabolic Optimization and Simulation Tool) is a software package that implements GDBB (Genetic Design through Branch and Bound) in an intuitive user-friendly interface with Excel-like editing functionality, as well as implementing FBA (Flux Balance Analysis), and supporting SBML (Systems Biology Markup Language) and CSV (Comma-Separated Values) files. GDBB is currently the fastest algorithm for finding gene knockouts predicted by FBA to increase production of desired products, but GDBB has only been available on a command line interface, which is difficult to use for those without programming knowledge, until the release of MOST.
Education
Boston University / Metropolitan College / Graduated: 2025
Master of Science – MS, Software Development
Ranked #8 in Online Information Technology Programs by US News
Taught by Computer Science faculty of Boston University with a focus on electives from BU MET Computer Science Master of Science. Online Program specializing on Enhancing Software Development skills. Focusing on Secure Coding, Software Engineering Methodologies, Data Science in Python, and Software Quality. As well as Data Structures and Algorithms, Software Designs and Patterns, Advanced Programming Techniques in Java. Languages explored include Python, Java, C, C++, and SQL.
Completed:
- MET CS 521 Information Structures with Python
- MET CS 526 Data Structures and Algorithms
- MET CS 622 Advanced Programming Techniques
- MET CS 665 Software Design and Patterns
- MET CS 673 Software Engineering
- MET CS 677 Data Science with Python
- MET CS 701 Rich Internet Application Development
- MET CS 763 Secure Software Development
Rutgers University/ Camden, NJ / Graduated: 2020
Bachelor of Science – Computer Science , Minor in Psychology
Member, Center for Computational and Integrative Biology Research Group
President, E-Commerce & IT Society
Software Development related electives
Computational Biology related Independent Studies
Main:
- Programming Fundamentals using Python (50:198:111)
- Object Oriented Programming using Python (50:198:113)
- C & Unix Programming (50:198:211)
- Senior Design Project using LAMP and Machine Learning (50:198:493)
- Data Structures (50:198:213)
- Design and Analysis of Algorithms (50:198:371)
- Operating Systems using C and Debian Linux with VMware (50:198:341)
Electives:
- Introduction to Computing for Engineers and Scientists using MATLAB (50:198:105)
- Software Engineering audited course using Java Web development and SQL (50:198:323)
- Java Applications (50:198:325)
- Database Systems using SQL (50:198:451)
- Cryptography (50:198:475)
Studies:
- Computational Biology using Java Desktop Development (50:198:494)
- Coarse Grain Simulation of Lipid Bilayer (50:750:490)
Highlighted Graduate Work
(Boston University CS 677 Final Project)
Relational data analysis and several machine learning algorithms. Creating a contingency table to find association between gene mutations to help visualize relations across gene mutation networks that were discovered in the Random Forest step. The Genes chosen to build the contingency table are non-deterministic per nature of Random Forest.